
Computer Science (CS)/Bioengineering (BE) M 182
This is a placeholder.
Computer Science (CS)/Computational & Systems Biology(C&SB)/Bioengineering (BE)/Ecology & Evolutionary Biology, CS-BE CM 186/286, EEB M 178

This combined undergraduate and graduate capstone course (M186 concurrently scheduled with course M286, in three departments) is normally offered in Fall Quarter only and includes four hours of lecture and a two hour simulation laboratory – for five credit units. An introductory course in signals and systems (e.g. EE 102) is co- or prerequisite.
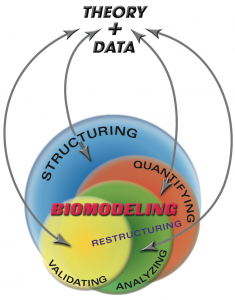
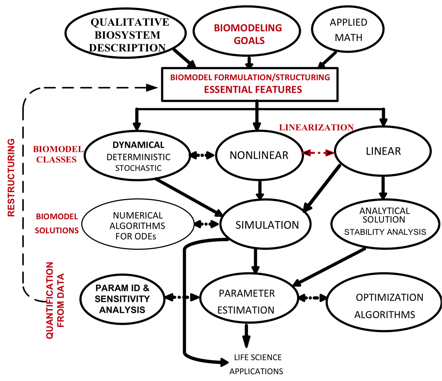
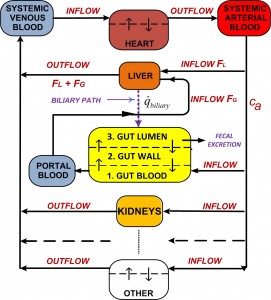
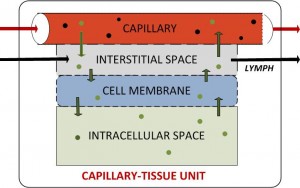
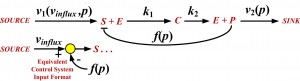
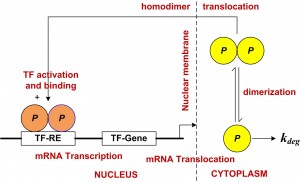
The course content covers the “how-to” and analytical aspects of dynamic biosystems modeling and computer simulation methodology for studying biological/biomedical/pharmaceutical processes and systems. The flowchart above depicts the topics covered and their interrelationships. Control system, multicompartmental, predator-prey, pharmacokinetic (PK), pharmacodynamic (PD) and other structural modeling methods are developed and applied to life sciences problems at multiple scales – molecular, cellular (biochemical pathways/networks), organ and organismic levels. The focus is primarily on data-driven modeling, translating biomodeling goals and data into mathematics models and implementing them for simulation and analysis. In this regard, the basics of numerical simulation algorithms are also covered, along with modeling software exercises in class and PC laboratory assignments for home and lab.
The 2015 textbook (Elsevier & Amazon) for course 186/286 – Systems Biology Modeling and Simulation – shown above on the right, consolidates and unifies classical and contemporary multiscale methodologies for math modeling and computer simulation of dynamic life science systems – from molecular/cellular, organ-system, on up to population levels. About 1/3 of the text is covered in the 186/286 course. The level is basic-to-intermediate, with much emphasis on biomodeling from real biodata, for use in real applications. The full textbook content is meant for a three quarter or two semester sequence of courses.
The book pedagogy is developed as a systematic tutorial, with clearly spelled-out and unified nomenclature – derived from the author’s own modeling efforts, publications and teaching over half a century – as well as numerous content and citations to from the basic modeling literature. One feature is that ambiguities in some concepts and tools in this literature are clarified and others are rendered more accessible and practical. Also featured are novel qualitative theory and methodologies for recognizing dynamical signatures in data using structural (multicompartmental and network) models and graph theory; and analyzing structural and measurement (data) models for quantification feasibility. Book topics include:
— The pertinent biology, biochemistry, biophysics or pharmacology for modeling are also provided, to support understanding the amalgam of “math modeling” with life sciences.
— Introductory coverage of core mathematical concepts such as linear and nonlinear differential and difference equations, Laplace transforms, linear algebra, probability, statistics and stochastics topics.
— Strong emphasis and material on quantifying as well as building and analyzing biomodels – especially from data. This includes methodology and computational tools for: parameter identifiability; sensitivity analysis; parameter estimation from real data; model distinguishability and simplification; and practical bioexperiment design and optimization.
— A companion website provides solutions and program code for examples and exercises using Matlab, Simulink, VisSim, SimBiology, SAAMII, Copasi and SBML-coded models: http://booksite.elsevier.com/9780124104112/downloads/PROGRAMS_CODE_FOR_CHAP_EXS_EXERCISES_4_22_14.pdf
OTHER COURSES
CS/C&SB/BE M184: Introduction to Computational and Systems Biology
This is a two hour survey/lecture course introducing students to the basics of computational and systems modeling and computation in biology and medicine, as well as research topics in these areas. It provides motivation, flavor, culture and cutting-edge contributions in computational biosciences and aiming for more informed basis for focused studies by students with computational and systems biology interests. In addition to some lectures on the basics, most lectures are by individual UCLA faculty researchers discussing their active computational and systems biology research. Requisites include some basic computer science knowledge (e.g. CS31 or PC10 A) and some basic math (e.g. Math 31A, 31B).
CS/C&SB/BE CM187/287: Research Communication in Computational and Systems Biology
This four hours-a-week, four-unit concurrently scheduled capstone II course is about completing, reporting, critiquing and finalizing computational and systems biology research in a publishable format. New projects are possible, but the primary goal is completion of thesis and publication level research projects already in progress. Major emphasis is on effective research reporting, both oral and written, via weekly oral presentations and written progress reports. Interactive discussion of research progress and written and oral reports in the classroom, with active participation of the all students as well as the instructor, is a key component of the pedagogy. Requisite: capstone course M186.