- Date: December 15, 2016
- Website
The p53 signaling system is one of the most important and studied control systems in cellular biology and its detailed structure is still very much in discovery mode. It’s known to be dysfunctional in most forms of cancer and this has been a major factor motivating numerous modeling efforts – generating a variety of different candidate structures as alternative mechanistic hypotheses – all directed toward better understanding of this system. Together, this variety of paradigms exemplify bottom-up modeling and remodeling well, for discovering what’s included, excluded and how components postulated to be included are interconnected. We’ve been working on discovering these major signaling pathways, based on data-driven modeling from real data collected in our collaborator Geofrey Wahl’s laboratory at the Salk Institute. This is all developed in Chapters 15 and 17 of Dynamic Systems Biology Modeling and Simulation (Author: Joseph DiStefano III), Academic Press/Elsevier, New York, Hardcover & Electronic Editions, 2015. The book is available at the URL above.
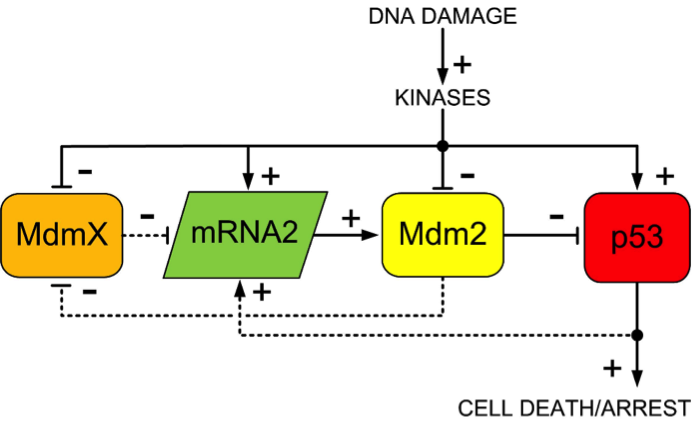
p53 regulation with MdmX included with the core regulator (Mdm2 with p53). Constitutive production and degradations ae excluded. Two signaling feedback loops are present: the (outer) negative one between p53 and Mdm2; and the (inner) positive one between MdmX and Mdm2.
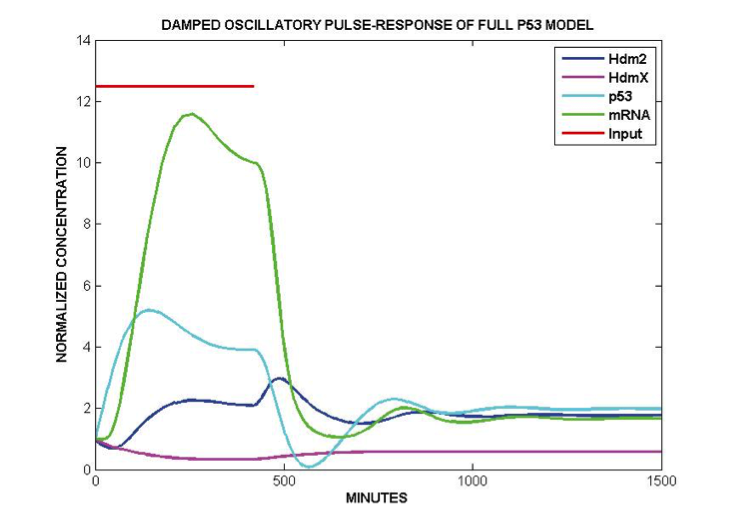
Simulated p53 signaling responses of our quantified model to a 420 minute pulse of kinase phosphorylation of p53. This model, quantified from real data, oscillates without the need for repeated input pulsing or time delays in the feedback.